SNPs (Single Nucleotide Polymorphisms) and Abasic Site
Single nucleotide polymorphisms (SNPs) are places along the chromosomes where the genetic code tends to vary from one person to another by just a single base. They are estimated to occur about once every 1000 bases along the 3-billion-base human genome. SNPs in genes or control regions may influence susceptibility to common diseases. Others probably have no function but could provide valuable markers for gene hunters; if they lie close to a susceptibility gene, they are likely to be inherited along with it.
DNA sequencing is the method of choice to identify SNPs. However, DNA sequencing is in most cases a cost- and time-consuming task. To overcome this, we propose here the use of an artificial DNA oligomer which contain an abasic site to detect SNPs. Abasic sites, where purine or pyridimine group is removed from a DNA strand and called as AP sites, are produced in mammalian cells upto 10,000 per day, and are also intermediates in the base repair pathways.
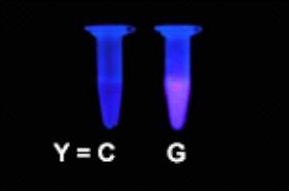
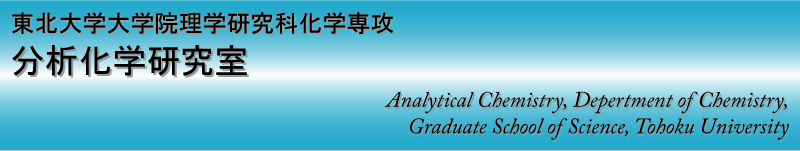
Fluorescence detection of the P177R
point mutation (C/G) in the p 53 gene
(5'-C TGC CYC CAC C-3' /
3'-G ACG GXG GTGG -5';
Y = C (normal) or G (mutation), X = AP site)
Small hydrogen bonding fluorescent receptor can be incorporated into a DNA oligomer containing an AP site and shows a change in fluorescence intensity, which enables us to differentiate point mutations easily with the naked eye. To illustrate the applicability of the technique to the screening of DNA mutations or polymorphisms, fluorescence detection of well-defined SNPs in the human p53 gene is demonstrated. The p53 protein is a checkpoint control of the cell cycle and induces cell growth arrest and apoptosis. Furthermore, alteration of the p53 gene is strongly involved in the promotion of tumors. Detection of genetic alterations in p53 is therefore clinically relevant for diagnosis and treatment of various types of tumors.
"Use of Abasic Site Containing DNA Strands for Nucleobase Recognition in Water"
K. Yoshimoto, S. Nishizawa, M. Minagawa and N. Teramae,
J. Am. Chem. Soc., 125(30), 8982-8983 (2003). [abstract]